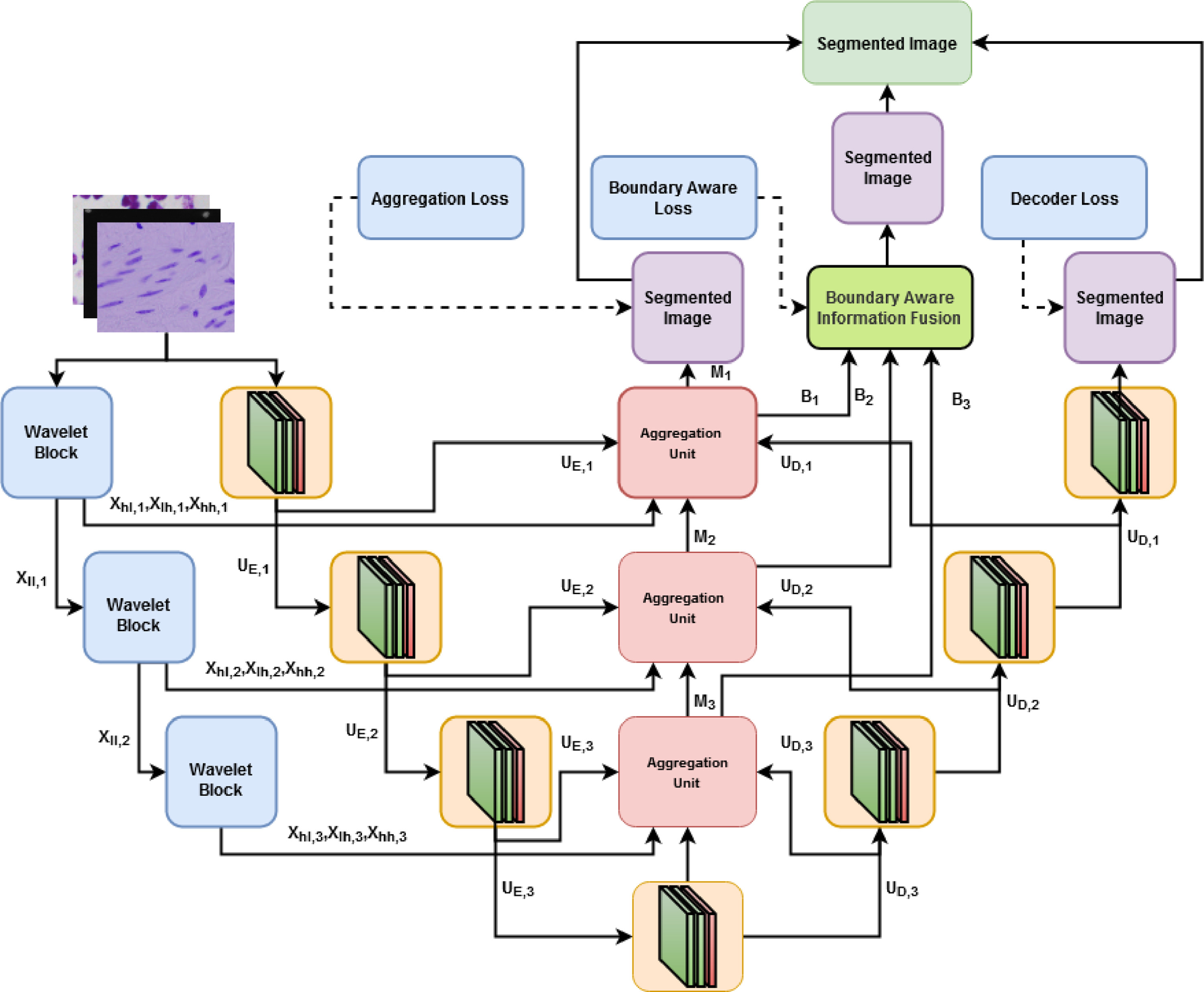
BAWGNet: Boundary aware wavelet guided network for the nuclei segmentation in histopathology images
Tamjid Imtiaz, SHaikh Anowarul Fattah; Sun-Yuan Kung
Aug 2023 - Computers in Biology and Medicine
Precise cell nucleus segmentation is very critical in many biologically related analyses and disease diagnoses. However, the variability in nuclei structure, color, and modalities of histopathology images make the automatic computer-aided nuclei segmentation task very difficult. Traditional encoder–decoder based deep learning schemes mainly utilize the spatial domain information that may limit the performance of recognizing small nuclei regions in subsequent downsampling operations. In this paper, a boundary aware wavelet guided network (BAWGNet) is proposed by incorporating a boundary aware unit along with an attention mechanism based on a wavelet domain guidance in each stage of the encoder–decoder output. Here the high-frequency 2 Dimensional discrete wavelet transform (2D-DWT) coefficients are utilized in the attention mechanism to guide the spatial information obtained from the encoder–decoder output stages to leverage the nuclei segmentation task. On the other hand, the boundary aware unit (BAU) captures the nuclei’s boundary information, ensuring accurate prediction of the nuclei pixels in the edge region. Furthermore, the preprocessing steps used in our methodology confirm the data’s uniformity by converting it to similar color statistics. Extensive experimentations conducted on three benchmark histopathology datasets (DSB, MoNuSeg and TNBC) exhibit the outstanding segmentation performance of the proposed method (with dice scores 90.82%, 85.74%, and 78.57%, respectively).
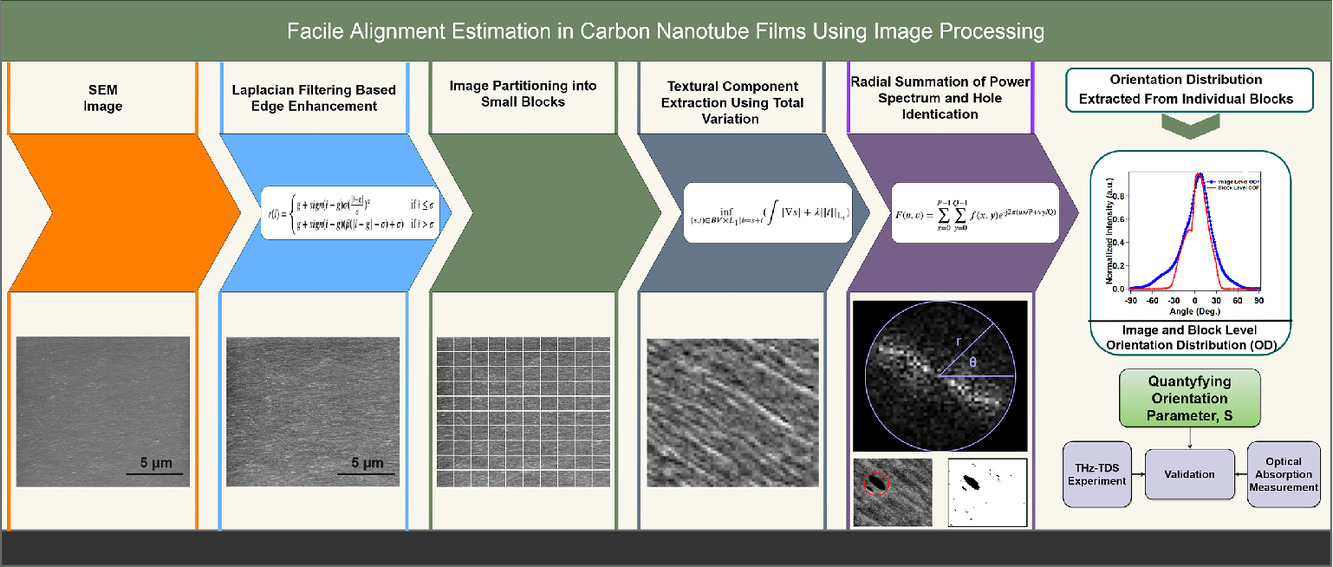
Facile alignment estimation in carbon nanotube films using image processing
Tamjid Imtiaz, Jacques Doumani, Fuyang Tay, Natsumi Komatsu, Stephen Marcon, Motonori Nakamura, Saunab Ghosh, Andrey Baydin, Junichiro Kono; Ahmed Zubair
Jan 2023 - Signal Processing
Whether a macroscopic assembly of carbon nanotubes can exhibit the one-dimensional properties expected from individual nanotubes critically depends on how well the nanotubes are aligned inside the assembly. Therefore, a simple and accurate method for assessing the degree of alignment is desired for the rapid characterization of carbon nanotube films and fibers. Here, we present an end-to-end solution for determining the global and local spatial orientation of carbon nanotubes in films within a short amount of time using a fast, precise, and economical approach based on an image processing method applied to scanning electron microscopy images. We first use Laplacian edge enhancement filtering for improving the appearance of edge regions, which is followed by image partitioning into multiple blocks to capture the nanoscale orientation characteristics and total variation-based image decomposition of these image blocks. We then perform a 2D-fast Fourier transform on the image decomposed textural components of these edge-enhanced image blocks to determine the orientation distribution, which is utilized to estimate the 2D nematic order parameter. To show the effectiveness of our method, we corroborated our results against results obtained with other state-of-the-art image processing and experimental techniques.
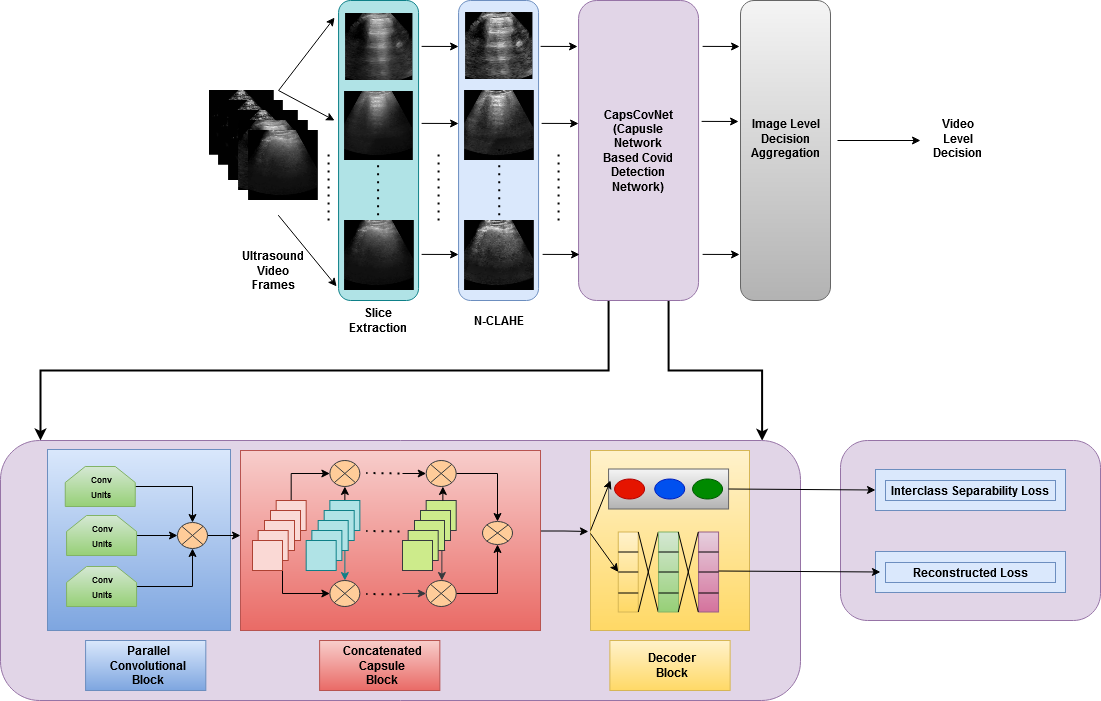
CapsCovNet: A Modified Capsule Network to Diagnose COVID-19 From Multimodal Medical Imaging
A F M Saif, T Imtiaz, S Rifat, C Shahnaz, W P Zhu; M O Ahmed
Aug 2021 - IEEE Transactions on Artificial Intelligence
Since the end of 2019, novel coronavirus disease (COVID-19) has brought about a plethora of unforeseen changes to the world as we know it. Despite our ceaseless fight against it, COVID-19 has claimed millions of lives, and the death toll exacerbated due to its extremely contagious and fast-spreading nature. To control the spread of this highly contagious disease, a rapid and accurate diagnosis can play a very crucial part. Motivated by this context, a parallelly concatenated convolutional block-based capsule network is proposed in this article as an efficient tool to diagnose the COVID-19 patients from multimodal medical images. Concatenation of deep convolutional blocks of different filter sizes allows us to integrate discriminative spatial features by simultaneously changing the receptive field and enhances the scalability of the model. Moreover, concatenation of capsule layers strengthens the model to learn more complex representation by presenting the information in a fine to coarser manner. The proposed model is evaluated on three benchmark datasets, in which two of them are chest radiograph datasets and the rest is an ultrasound imaging dataset. The architecture that we have proposed through extensive analysis and reasoning achieved outstanding performance in COVID-19 detection task, which signifies the potentiality of the proposed model.
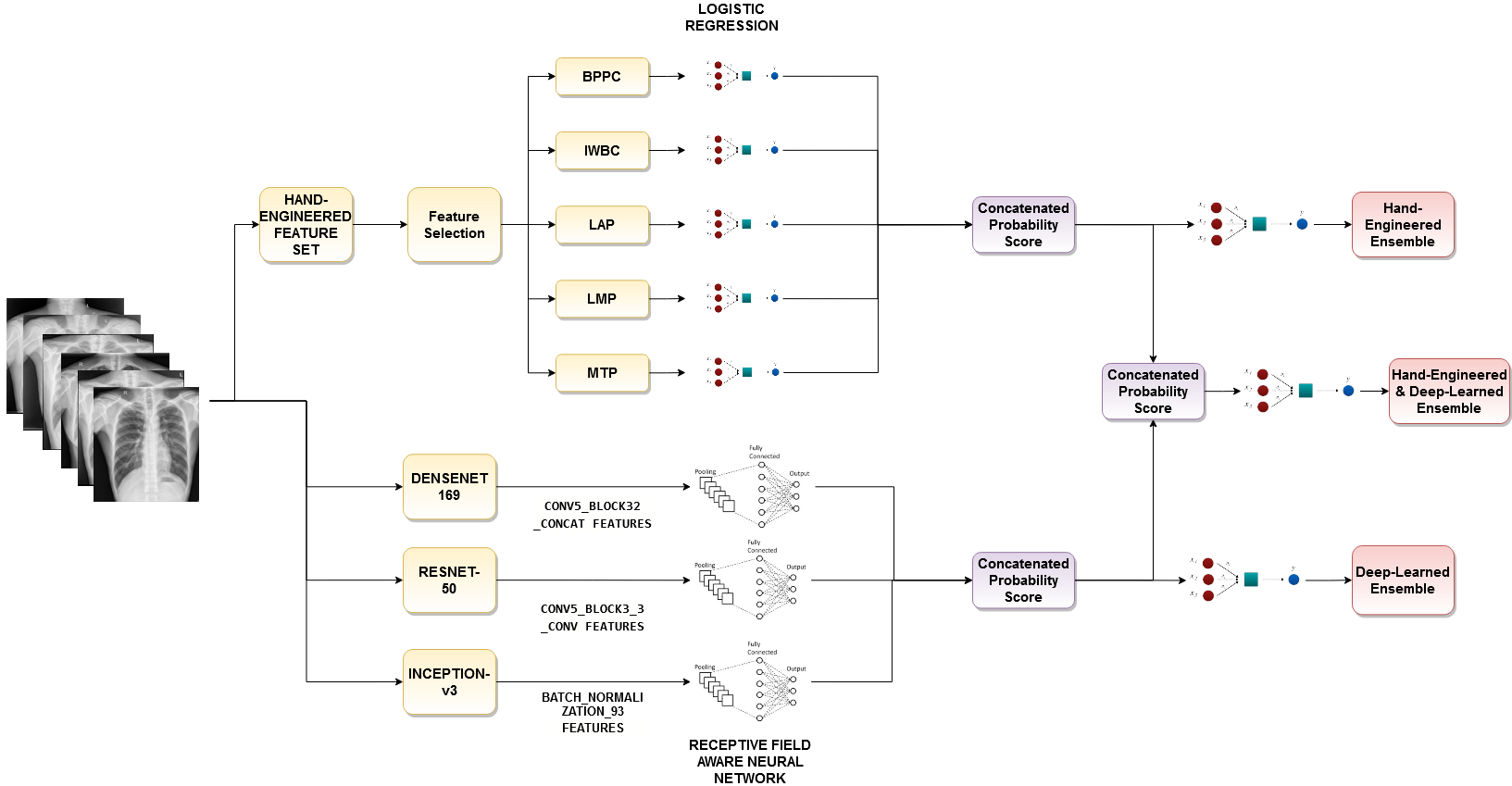
Exploiting Cascaded Ensemble of Features for the Detection of Tuberculosis Using Chest Radiographs
A F M Saif, T Imtiaz, C Shahnaz, W P Zhu; M O Ahmed
Aug 2021 - IEEE Access
Tuberculosis (TB) is a communicable disease that is one of the top 10 causes of death worldwide according to the World Health Organization. Hence, Early detection of Tuberculosis is an important task to save millions of lives from this life threatening disease. For diagnosing TB from chest X-Ray, different handcrafted features were utilized previously and they provided high accuracy even in a small dataset. However, at present, deep learning (DL) gains popularity in many computer vision tasks because of their better performance in comparison to the traditional manual feature extraction based machine learning approaches and Tuberculosis detection task is not an exception. Considering all these facts, a cascaded ensembling method is proposed that combines both the hand-engineered and the deep learning-based features for the Tuberculosis detection task. To make the proposed model more generalized, rotation-invariant augmentation techniques are introduced which is found very effective in this task. By using the proposed method, outstanding performance is achieved through extensive simulation on two benchmark datasets (99.7% and 98.4% accuracy on Shenzhen and Montgomery County datasets respectively) that verifies the effectiveness of the method.
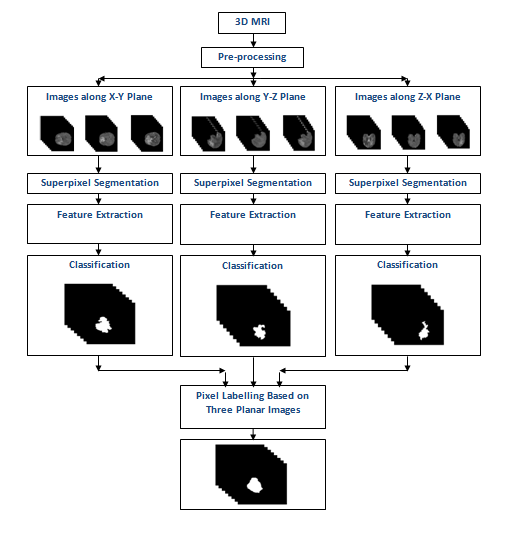
Automated Brain Tumor Segmentation Based on Multi-Planar Superpixel Level Features Extracted From 3D MR Images
T Imtiaz, S Rifat, S A Fattah; K A Wahid
Dec 2019 - IEEE Access
Brain tumor segmentation from Magnetic Resonance Imaging (MRI) is of great importance for better tumor diagnosis, growth rate prediction and radiotherapy planning. But this task is extremely challenging due to intrinsically heterogeneous tumor appearance, the presence of severe partial volume effect and ambiguous tumor boundaries. In this work, a unique approach of tumor segmentation is introduced based on superpixel level features extracted from all three planes (x -y, y - z, and z - x) of 3D volumetric MR images. In order to avoid the pixel randomness and to account for precise inhomogeneous boundaries of brain tumor, each of the images belonging to a particular plane is partitioned into irregular patches (superpixels) based on their intensity and spatial similarity. Next, various statistical and textural features are extracted from each superpixel where all three planes are considered separately in order to obtain better labeling on superpixels in tumor edges. A feature selection scheme is proposed based on their performance on histogram based consistency analysis and local descriptor pattern analysis, which offers a significant reduction in feature dimension without sacrificing classification performance. For the purpose of supervised classification, Extremely Randomized Trees is used to classify these superpixels into a tumor or a non-tumor class. Finally, pixel level decision is taken based on corresponding decisions obtained in each plane. Extensive simulations are carried out on publicly available dataset and it is found that the proposed method offers better tumor segmentation performance in comparison to that obtained by some state of the art methods.





